Casimir Effect
A consensus is forming that Casimir effect, which is a macroscopic manifestation of quantum fluctuations of vacuum, is instrumental in nano-mechanical devices in terms of both desirable and undesirable effects. This means that a comprehensive understanding of the interactions between dielectric, metallic, and magnetic objects of arbitrary geometry is needed. This, however, is a nontrivial task.
We have recently developed a perturbative theoretical scheme that can be used for such calculations, and used it to study a number of systems including dielectric hetero-structures and meta-materials.
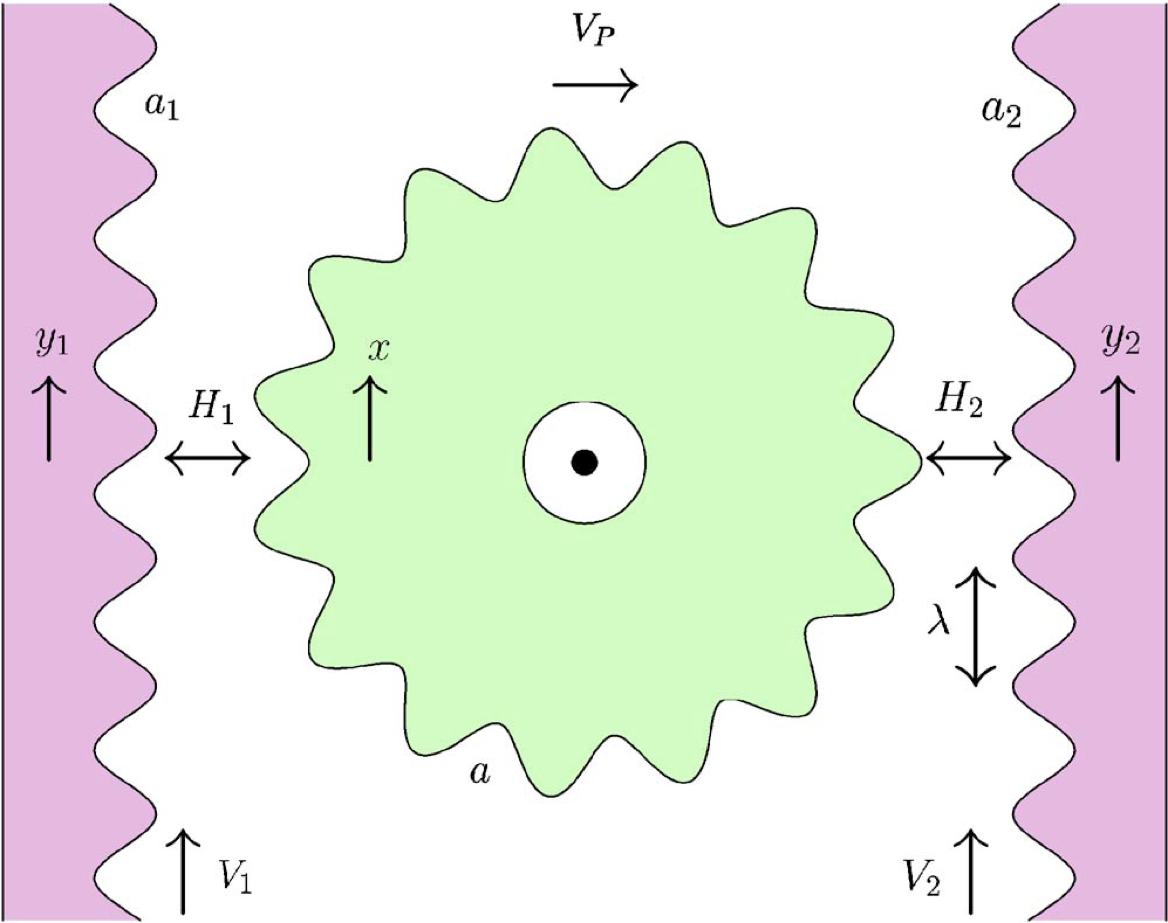
We have also been working on a number of proposed nano-mechanical devices that could use the lateral Casimir force between corrugated surfaces for non-contact friction-less mechanical coupling. The force transduction between two surfaces via the quantum vacuum is realised by a term proportional to the sinus of the phase difference between them, which is reminiscent of the Josephson coupling between superconductors. Because the coupling is nonlinear, its mechanical response could involve oscillatory and unstable behaviours. For a number of such devices, we have studied the dynamical phase diagram of the system in the space of relevant parameters and determined the feasibility of such non-contact ratchets and gears, their response to external load, as well as their mechanical efficiency.
Group members involved: R. Golestanian
DNA Elasticity
Mechanical properties of DNA have been subject of intense recent interest with the advent of manipulation experiments. An elastic model has been developed to study these properties, with the practical advantage that it reduces the large number of atomistic degrees of freedom to just 6 per base-pair. This coarse-grained elastic model, equipped with a rough average estimate for a few key elastic constants such as average bending modulus, twist modulus, and stretch modulus, has successfully accounted for micron-scale DNA manipulation experiments. We have shown that such simple elastic models are valid even at relatively small scales and high curvatures, using a detailed comparison with recent high precision X-ray structure of bent DNA inside the nucleosome complex. Part of the success in this description comes from considering the anisotropy in the bending rigidity of DNA at the base-pair level, which could predict nonlinearities in the shape of short DNA loops in the form of kinks.
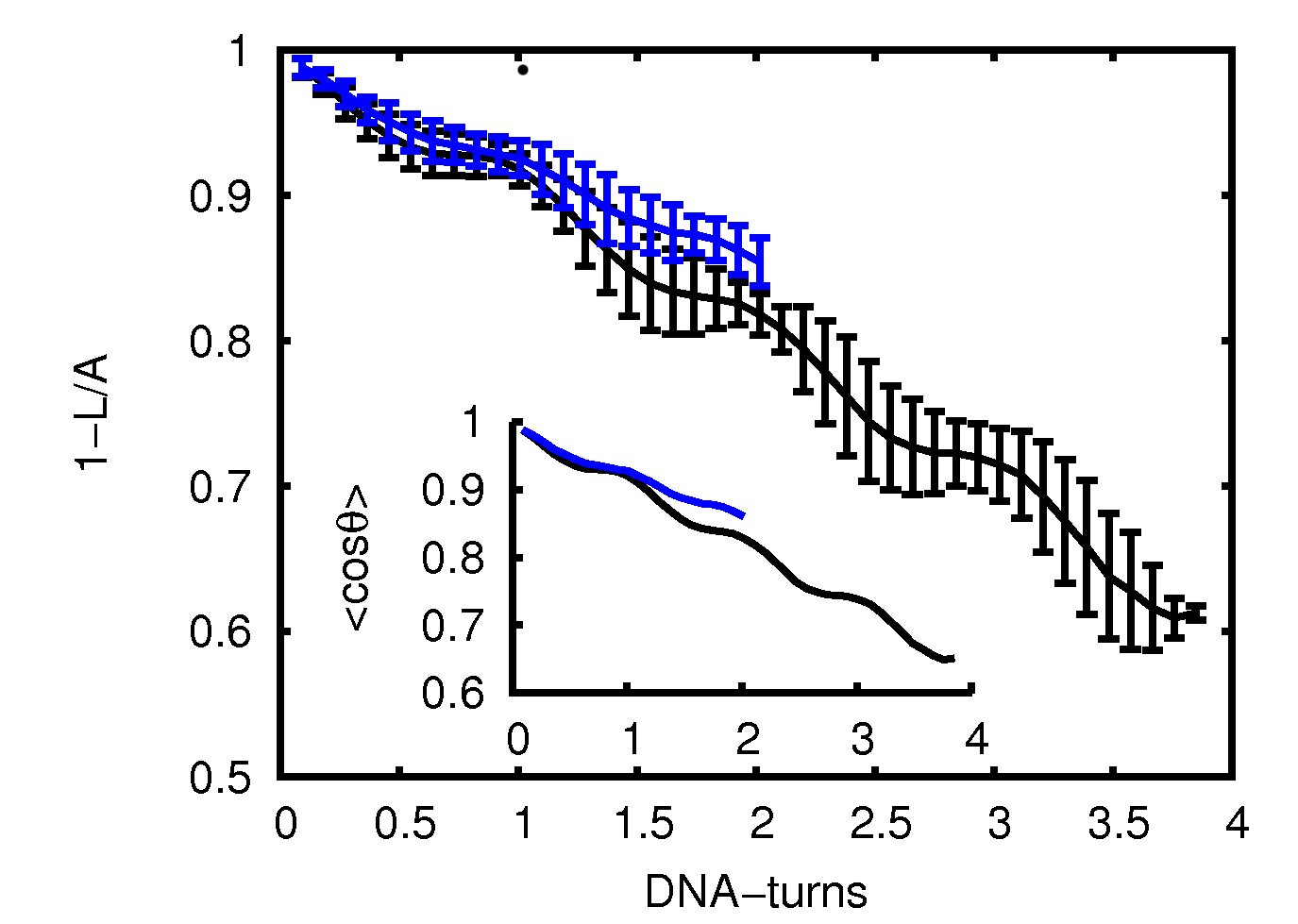
That is why we have turned to the atomistic molecular dynamics simulations as a very useful technique which offers potentially the most spatially detailed descriptions of the DNA elasticity. Firstly, we analyzed the dinucleotide energy landscape carefully. In particular, we evaluated the DNA elasticity cross-terms demonstrating DNA asymmetry at very short scale is more complex than predicted by long-scale DNA models, with the cross-terms relating torsion, bending, lateral shear and stretch modulus being most essential. Recently, by probing the conformation and fluctuations of DNA from single base-pair level up to four helical turns, we have examined the potential effect of the asymmetric double-helical structure on the elastic response of the molecule to find out at which scale DNA starts to behave as an isotropic elastic rod with the elastic constants approaching the large-scale experimental values [4]. We have found evidence that supports cooperative softening of the stretch modulus and identify the essential modes that give rise to this effect (Fig. 1 and Movie 1 and 2). The bend correlation exhibits modulations that reflect the helical periodicity, while it yields a reasonable value for the effective persistence length (Fig. 2), and the twist modulus undergoes a smooth crossover—from a relatively smaller value at the single base-pair level to the bulk value—over half a DNA-turn.
The success of these approaches suggests that an improved description that uses sequence dependent elastic moduli may help us address biologically relevant mechanical properties of DNA during complexation with proteins (which happen at nano-scale) and could in turn shed some light on dynamical properties of DNA during transcription. We aim to combine the two formulations into a multi-scale description of DNA elasticity that could be used at the intermediate scale to study DNA protein interactions.
Group members involved: A. Noy (alumna), R. Golestanian








