Bias in the the arrival of variation
Nothing in Biology Makes Sense Except in the Light of Evolution."
wrote the great naturalist Theodosius
Dobzhansky, but to really understand evolution, a stochastic
optimization process in extremely high dimensional spaces[1], will require
techniques from statistical physics.
Darwinian evolution proceeds by two steps. First, random mutations generate new heritable phenotypic variation. Second, the process of natural selection ensures that phenotypes with higher fitness are more likely to dominate in a population over time. Research in evolutionary theory has mainly focussed the second step, natural selection. Much less is known about the first step, the arrival of variation. For a popular introduction to this topic,
see
The Arrival of the Fittest by Andreas Wagner. There is also an excellent Wikipedia article entitled Bias in the introduction of variation.
Much of our work in this area uses theoretical tools of statistical mechanics and
algorithmic information theory [2], together with computer simulations,
to study genotype-phenotype maps, which give access to the structured role of variation in evolution.
As an example, in this paper on phenotypic bias in functional RNA [3], we are able to quantitatively predict the frequencies with which RNA structures appear in nature, without taking
natural selection into account. Instead, a very strong bias in the arrival of variation dominates through a non-ergodic effect we call the arrival of the frequent [4].
To illustrate how extremely strong such biases in the introduction of new variation can be, consider L=126 length RNA,
where making every possible strand would exhaust all the observable matter in the universe. In spite of this hyperastronmically large [1] space of possibilities, we find that
only 68 out of 1012 structures are used in nature , and that these are all within the 100 most likely structures to appear upon random sampling of sequences. The vast majority of biophysically possible structures are not used at all.
Such a dramatic effect raises a big question: does this such developmental bias also play a
key role in determining the evolutionary outcomes for other biological GP maps, including larger-scale developmental systems?
In another recent paper [5] we show that this bias towards simplicity also implies a bias towards structures with high symmetry which we observe in protein quaternary structures.
The big challenge for this postdoc project is test whether such patterns can be observed beyond these molecular phenotypes, at the larger scales of the evolution of development.
Deep and general arguments based the coding theory from algorithmic information theory [2,5] suggest that strong bias
may hold also for some aspects of development, but that hypothesis first needs to be established (or falsified) explicitly by detailed calculations and comparison to experimental reality.
In this project, you will use a wide range of techniques to explore the mapping from genotypes to phenotypes in models of development,
and to study the adaptive evolutionary dynamics on these landscapes[6].
There are close analogies to the question of why overparameterised deep neural networks generalize so well [7],
and part of the project may include an exploration of these commonalities.
This is an ambitious and challenging interdisciplinary project.
Strong quantitative skills and a proven track record of creative and successful independent research are the most important qualities we are looking for. Experience with biological physics is a plus, but not a requirement.
[1]
Contingency, convergence and hyper-astronomical numbers in biological evolution
Ard A. Louis
Studies in History and Philosophy of Biological and Biomedical Sciences 58, 107 (2016)
[2]
Input–output maps are strongly biased towards simple outputs
K. Dingle, C. Q. Camargo and A. A. Louis
Nature Comm. 9, 761 (2018)
[3]
Phenotype bias determines how RNA structures occupy the morphospace of all possible shapes
Kamaludin Dingle, Fatme Ghaddar, Petr Sulc, and Ard A. Louis
Molecular Biology and Evolution, 39, msab280 (2021)
|
biorxiv abstract
[4]
The arrival of the frequent: how bias in genotype-phenotype maps can steer populations to local optima
Steffen Schaper and Ard A. Louis
PLoS ONE 9(2): e86635 (2014)
| arxiv abstract
|
[5]
Symmetry and simplicity spontaneously emerge from the algorithmic nature of evolution
Iain G Johnston, Kamaludin Dingle, Sam F. Greenbury, Chico Q. Camargo, Jonathan P. K. Doye, Sebastian E. Ahnert, Ard A. Louis
PNAS 119, e2113883119 (2022).
|
biorxiv abstract
[6]
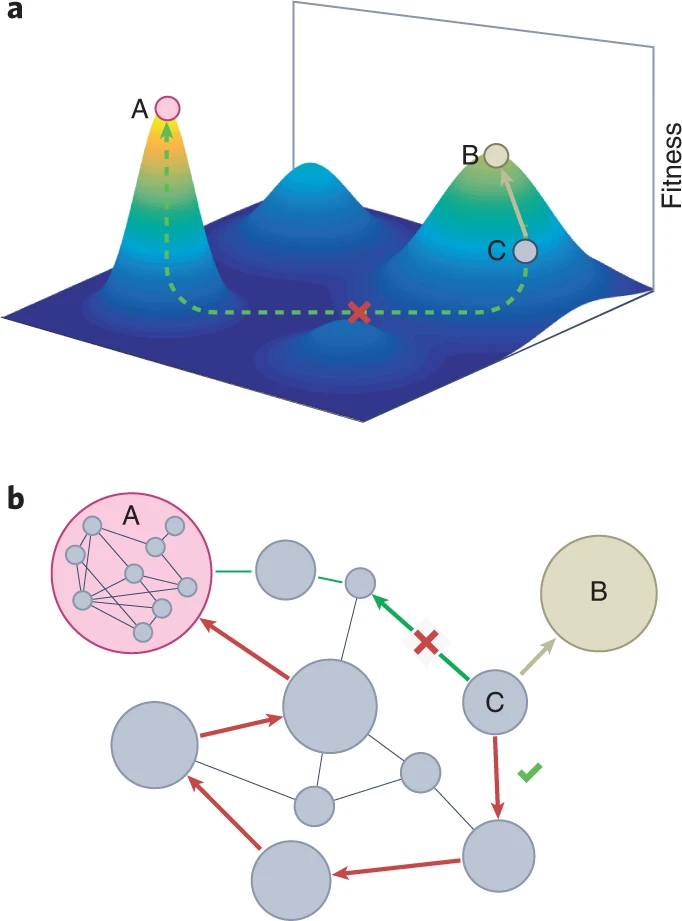
The structure of genotype-phenotype maps makes fitness landscapes navigable
Sam F. Greenbury, Ard A. Louis, and Sebastian E. Ahnert
Nature Ecology and Evolution (2022)
|
biorxiv abstract |
pdf |github repository|
|Nature Ecology Evolution Commentary
 |
|
[7]
Deep learning generalizes because the parameter-function map is biased towards simple functions
Guillermo Valle Pérez, Chico Q. Camargo, Ard A. Louis
| arxiv abstract
Further resources
Popularisations of our recent work
|
